SILAC Labeling Quantitative Proteomics
Introduction:
This service is designed for the quantitative and qualitative analysis of cell based systems (SILAC cell lines) or animal based systems (SILAC mice or rats). The results from this experiment provide a catalogue of the proteins present in all samples and a statistical analysis reflecting the changes in the protein levels observed across the samples (treated vs control), along with associated pathway information analyzed through IPA.
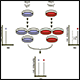
Stable isotope labeling with amino acids in cell culture (SILAC) is a quantitative proteomics technique that involves the incorporation of a label into proteins in vitro or in vivo prior to analysis by LC/MS/MS. Cells are labeled during the culturing process using media containing light or heavy amino acids, animals (eg, mice) are fed with the food containing light or heavy amino acids, the heavy amino acids have stable isotope atoms incorporated e.g. 13C6,15N2-Lysine or 13C6,15N4-Arginine. The labeled amino acids are used in protein synthesis, after several passages for cell lines and after three to four generations for mice, the labeled residues have been fully incorporated into the proteins. The labeled amino acids (heavy) are equivalent to their unlabeled counterparts (light) and their presence has no impact on the biological systems of SILAC cell lines, or the development, growth, and behavior of SILAC animals.
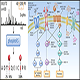
Cells or tissue lysates from light and heavy cultures can be lysed and mixed to create a population of both heavy and light labeled pairs of proteins (could be as many as 10 - 20 channels). The pooled sample is then processed through our general proteomics sample preparation pipeline. The number of fractionations dictates the depth of coverage and the time to analyze a single sample. Protein differential expression is calculated as a function of the peak areas of the light and heavy peptides detected in the sample. One single SILAC project can accommodate and quantify as many as ten to twenty groups of samples simutanously.
There are typically 10 steps in a SILAC labeling Omics Technologies quantitative proteomics project:
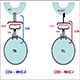
1) Cell culture – Cell lines (treated and untreated) were cultured with required culture media that are identical except in one respect: the first media contains the ‘light' and the other a ‘heavy' form of a particular amino acid (for e.g. 13C6,15N2-Lysine or 13C6,15N4-Arginine).
2) Sample preparation – Cell lysates collected; protein quantification on all samples.
3) Sample digestion – Reduction and alkylation followed by in-solution trypsin digestion and C18 cleaning before labeling.
4) Sample pooling – Equal amounts (according to total protein quantification) of labeled samples are pooled together to allow unbiased downstream purification and Mass Spectrometry Analysis.
5) SCX cleaning – Ion-exchange to remove excessive chemicals and fractionation of pooled sample through off-line SCX HPLC.
6) 2D fractionation – bRPLC separation of the SCX cleaned samples into 96 fractions, and further pooled into 24 fractions according to MyProt™-iPooling algorithm (based on peptide hydrophobicity, IEP, etc.)
7) Nano LC/MS/MS – 50 nL/min liquid chromatography to enable super high ionization efficiency for downstream Mass Spectrometry Analysis using the state-of-the-art Orbitrap Mass Spectrometer. 24 fractions will be analyzed in 24 Mass Spectrometry runs.
8) Protein profiling – Protein database searching with Mascot, Sequest HT, PEAKS de novo approaches, etc.
9) Data analysis – Data validation, visualization and quantification using commercial softwares and Omics Technologies's unique R packages and scripts.
10) Report generation – Report will be sent to you in Excel format as well as a summary in PDF format. We will also provide you any details you need for your papers' MATERIALS AND METHODS section. We will make sure you understand your result and help you with your paper writing with free follow-up services.
Sample types we accept:
1, Live cell lines or genetically engineered cell lines.
2, Cell Lysates and Tissue Lysates
3, Customized sample types (please contact us to discuss)
Please refer to our Omics Technologies Proteomics Sample Preparation and Shipping Guidelines for instructions on how to prepare your samples with less contamination for Mass Spectrometry Analysis.
-
Phone:
+1 888-206-0066 (TOLL FREE) -
Email:
info@omicstech.com